Our research
Our work lies at the interface of chemistry, biophysics, and computer science. Here are some research themes of current interest.
Identifying hidden alternative conformations of proteins and ligands in biophysical data
We study proteins as conformational ensembles. Although X-ray crystallography is an ensemble experiment, the results are typically summarized with a single static structure. We develop software to discover the structural ensembles present in the crystal (or on the EM grid). The ensemble nature of proteins highlighted by this work feeds into all of our mechanistic studies that interpret the functional effects of mutations, that characterize designed and artificially-evolved proteins, or that seek to modulate protein function with small molecules. These methods development efforts are central to discovering new allosteric ligands, through high throughput crystallographic fragment screening efforts. We are expanding, this direction to include modeling and validating protein structural data generated by cryo-electron microscopy (using EMRinger and ensemble modeling) and through integrative approaches to discover cryptic ligand binding sites. These methods development efforts are central to discovering new allosteric ligands, through high throughput crystallographic fragment screening efforts.
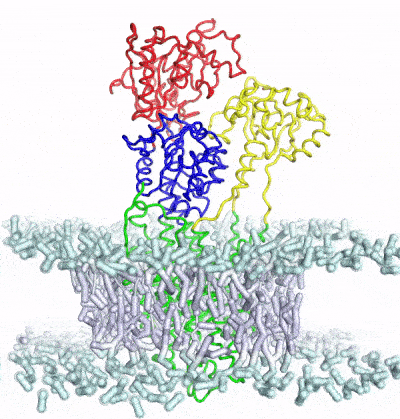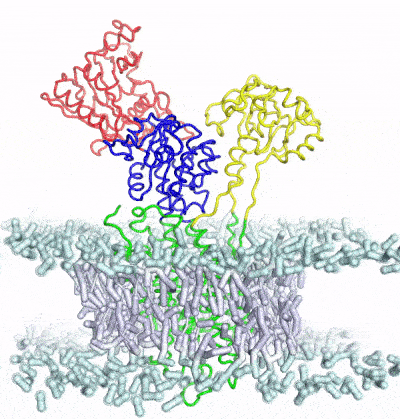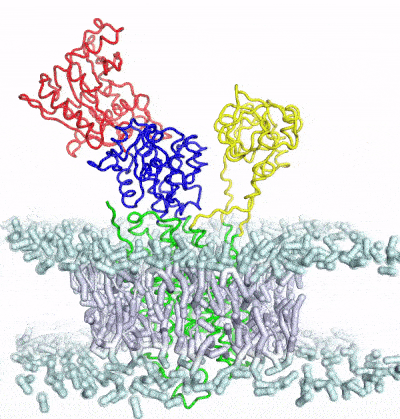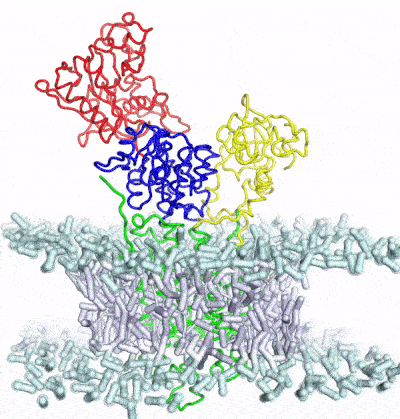
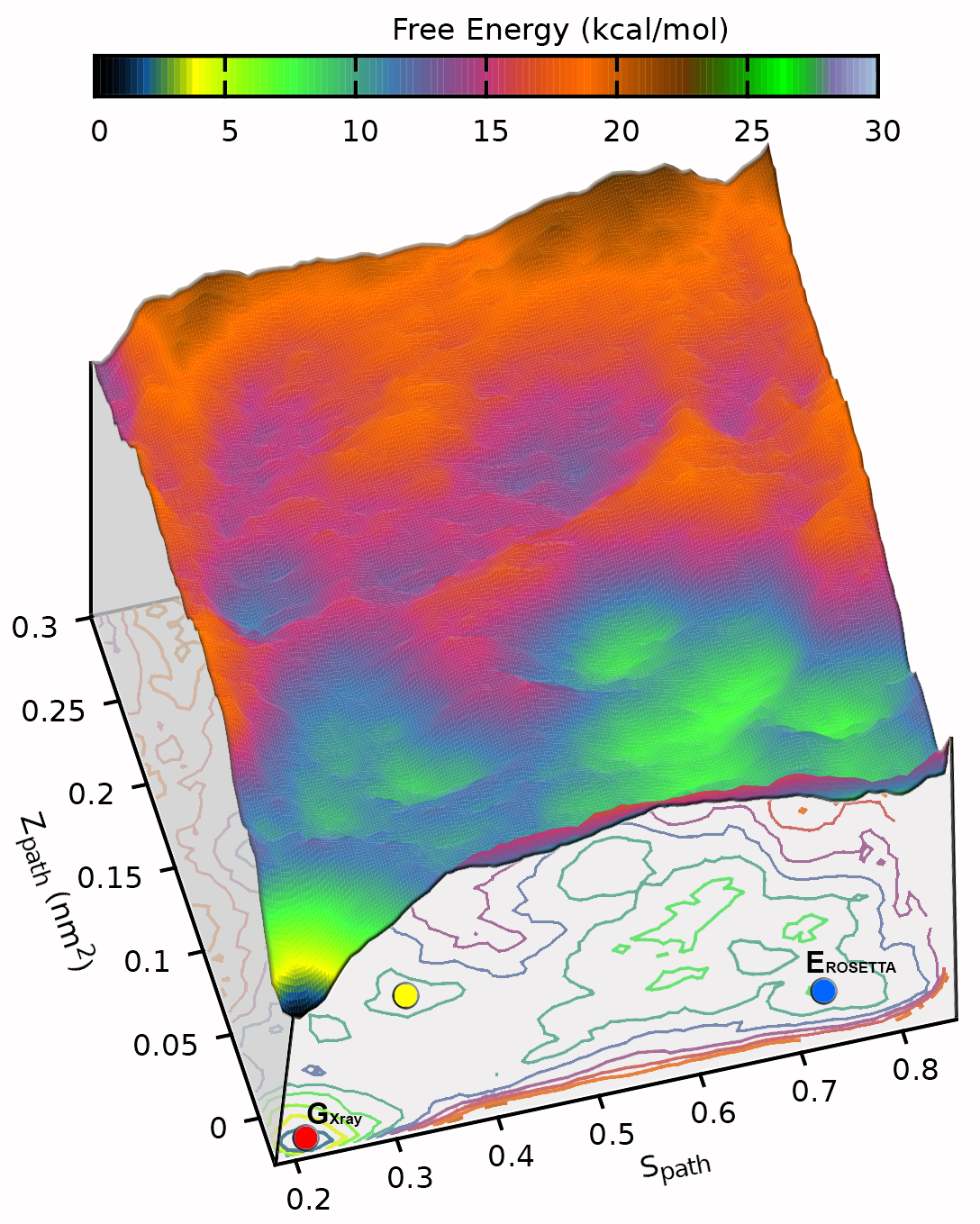
Develop and apply enhanced sampling techniques to extract thermodynamic and kinetic information of biomolecular dynamics
A major limitation of most biophysical techniques is the inability to directly reveal correlations in motions between distinct regions of macromolecules. We are taking advantage of the new capabilities of next-generation X-ray free electron laser (X-FEL) light sources to perform radiation damage-free imaging of proteins and to watch how protein ensembles respond when perturbed by rapid temperature jumps using the X-FEL. At equilibrium, X-ray diffuse scattering has the potential to reveal these motions; however, we currently lack the ability to collect, integrate, and refine diffuse scattering data. Our long-term goal is to increase the information content of every X-ray diffraction experiment to reveal atomic level coupling at high resolution and improved models of grouped flexibility at low resolution.
Integrative structual biology by combining multi-scale modeling with experimental data with the aid of enhanced sampling
A major limitation of most biophysical techniques is the inability to directly reveal correlations in motions between distinct regions of macromolecules. We are taking advantage of the new capabilities of next-generation X-ray free electron laser (X-FEL) light sources to perform radiation damage-free imaging of proteins and to watch how protein ensembles respond when perturbed by rapid temperature jumps using the X-FEL. At equilibrium, X-ray diffuse scattering has the potential to reveal these motions; however, we currently lack the ability to collect, integrate, and refine diffuse scattering data. Our long-term goal is to increase the information content of every X-ray diffraction experiment to reveal atomic level coupling at high resolution and improved models of grouped flexibility at low resolution.
